Supercharging Antibody Drug Discovery with AI: The Rise of Chai-2 and Patsnap’s Lao Tzu Dataset
Overview
In the fast-evolving world of biotech, artificial intelligence (AI) is reshaping how we discover and develop therapeutic antibodies. Echoing BBC’s recent report on the role of artificial intelligence and antibiotic design
AI can enable us to come up with molecules, cheaply and quickly, further expanding on our existing arsenal, hence giving us a leg up in the battle of our wits against the genes of superbugs.
If AI can help us design new antibiotics to fight resistant bacteria, imagine what it can do for antibodies. Antibody developers face the same challenge: discovering effective molecules quickly, at scale, and with precision. That’s where new generative models like Chai-2, powered by high-quality data, come in.
In August 2025, Chai Discovery raised $70 million Series A round to advance its AI platform for molecular design – a clear signal of the growing momentum and investor confidence in this space. At the forefront of this transformation is Chai-2, a generative diffusion model introduced by Chai Discovery with support from OpenAI. This breakthrough model marks a significant leap in de novo antibody design, enabling the generation of protein sequences with stable 3D structures — without relying on traditional databases or high-throughput screening yet still capable of increasing speed-to-clinic with confidence.
Why Chai-2 Matters
Chai-2’s impact in antibody discovery is profound:
- 100x improvement in success rate over conventional methods
- Design time reduced by over 50%, from several months to just under 2 weeks
- End-to-end generation of therapeutic molecules with functional binding sites
By integrating antigen-specific guidance into its full-atom diffusion framework, Chai-2 can generate antibodies tailored to specific epitopes — streamlining the drug discovery process and opening doors to previously unattainable therapeutic possibilities.
The Power of Diffusion Models in Antibody Design
Diffusion models have emerged as a dominant force in generative biology. Their ability to transform random noise into meaningful protein structures allows for:
- High-quality, diverse antibody candidates
- Scalable and repeatable design processes
- Exploration of vast sequence spaces for optimal variants
From AbDiffuser to AntiBARTy and RFdiffusion, a growing ecosystem of diffusion models is being trained on curated datasets to push the boundaries of antibody engineering.
Challenges in Antibody Development
Despite these advancements, AI-driven antibody design faces hurdles:
- Accessibility of complete, non-fragmented and/or unbiased public datasets to train and validate AI model
- Lack of negative training data can skew results
- Important biochemical properties like solubility and immunogenicity are often overlooked
Without better training foundations, AI risks generating antibodies that look good in silico but fail in the lab.
How Patsnap Can Help You
To address these, Patsnap has developed the Lao Tzu Antibody-Antigen Dataset — a meticulously curated repository combining AI-driven analysis with expert manual annotation. With over:
- 120,000+ antibody-antigen pairings
- 3,300 target antigens
- 20,000 affinity measurements
- 24,000 IC50/EC50 datapoints
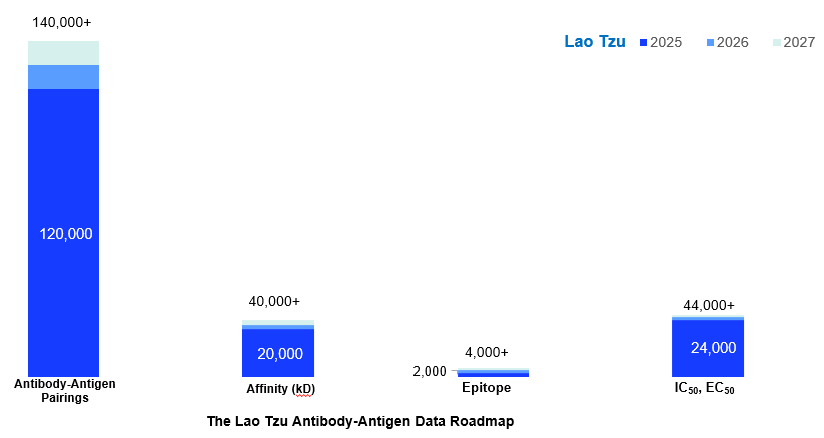
From trusted, patent and non-patent sources, this dataset empowers researchers to train and validate generative models with unprecedented accuracy and efficiency, identifying and developing therapeutic antibody with confidence.
Unlock the Antibody Dataset Mini Report and Learn How Quality Data Drives Better Research
Looking Ahead
As diffusion-based models continue to evolve, integrating novel algorithmic frameworks and leveraging large language models will further enhance their capabilities. Patsnap’s commitment to expanding the Lao Tzu dataset ensures that innovators have access to the high-quality data needed to drive the next wave of breakthroughs in antibody and protein design.
Stay ahead of the game and see how Patsnap’s Lao Tzu dataset can help supercharge your innovation efforts in driving antibody and protein design.